SEGMENTATION OF LUNG CT IMAGES AND NODULE DETECTION WITH MATLAB
SEGMENTATION OF LUNG CT IMAGES AND NODULE DETECTION WITH MATLAB
Segmentation of Lung CT Images and Nodule Detection with MATLAB
In this study, segmentation of lung CT images from medical images is described.
Segmentation is the first and most important step in image processing. The high error in segmentation will also affect the subsequent image processing stages, especially in biomedical images. For this reason, it is very important to make the segmentation with the least mistake.
-According to the discontinuities in the gray level values, the basic approach in the image segmentation problem is to divide a view based on the sudden changes in the gray levels which is against edge detection.
-Image segmentation according to similarities in gray level values is called region segmentation and is based on thresholding, growing, and split-and-merge operations. Based on similarities or differences in the gray level values of the pixels, an image segmentation concepts can be applied to both static and dynamic (time varying) images.
Matlab-based software codes have been used to reduce the number of parasites in the image, to detect the nodule as a cancerous structure in the lung, and to eliminate the lung organ from the image.
CT image properties of in this project:
422x511 uint8
Lung image with cancer cells (.jpg)
 |
| CT Image |
To get matlab image with imread command,
To spread the pixels in the image with the imopen command,
Using the region growing function to select the region in the image:
image=imread('CT_2.JPG');
image=rgb2gray(image);
se = strel('disk',2);
image = imopen(image,se);
figure, imshow(image,[]), hold all
%regiongrowing
%Inputs:
% cIM: 2D/3D grayscale matrix {current image}
% initPos: Coordinates for initial seed
position {ginput position}
% thresVal: Absolute threshold level to be
included {5% of max-min}
% maxDist: Maximum distance to the initial
position in [px] {Inf}
% tfMean: Updates the initial value to the
region mean (slow) {false}
% tfFillHoles: Fills enclosed holes in the
binary mask {true}
% tfSimplify: Reduces the number of vertices
{true, if dpsimplify exists}
poly = regiongrowing(image, [],
50, 150, false, true, true);
.
.
.
imshow(image,[]);
hold on;
plot(poly(:,1), poly(:,2), 'LineWidth', 1)
title('Region Of
Growing Segmented');
 |
| Region Selection |
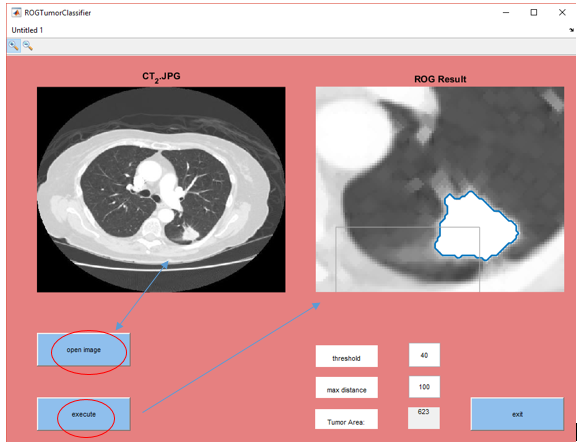
I can select with the cursor and select the tumor region.
The imopen command was used to precisely handle color transitions.
 |
| Tumor Area |
GUI:
We enter the threshold value and we set max distance.
The tumor area is calculated by matlab.
*When we changed of threshold value:
Area was calculated 582.
I get the best result
when we enter the correct threshold values and the correct distance values to
determine the region.
Results:
In the thesis, according to the image on hand, first the erosion process is followed by the dilation process or the first dilation process followed by the erosion process.
Both of these processes must be used at the same time otherwise the image will be distorted too much and we will not get the corrct result.
The pixel has been changed to correctly to select the region.
Then region growing applications were made and the region to be implemented was selected.
I applied the imopen command against the fisrt applications in the thesis. so I got a good result and a shorter process.
If imopen is not applied, a region with the wrong accuracy rate is selected.
I can select the damaged region with the cursor, i can calculate the size with the help of matlab.
Then i can obtain information about the tumor by entering appropriate values in patient controls.
Görüntü İşleme
M.K.
More information about of thesis:



Yorumlar
Yorum Gönder